Our research involves the development of computer programs that are used in all stages of cryo-EM projects — from sample preparation, data collection, image analysis and 3D reconstruction, structural analysis, to training. The following programs are distributed for free academic use.
CryoVR
Virtual reality-augmented hands-on cryo-EM training
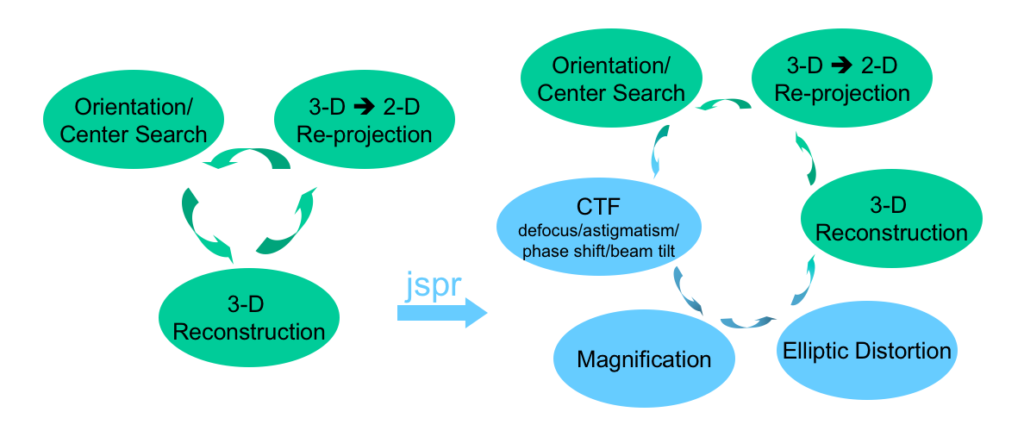
jspr
Software for single particle cryo-EM image processing and 3-D reconstruction. It is built on top of EMAN and EMAN2. Publication | Software
HLM
Classification of helical polymers with deep-learning language models. Publication | Software
HILL Web App
HI3D Web App
HelicalLattice Web App
Site 1 (click me) | Site 2 (click me)
map2seq Web App
ProCart Web App
CTF Simulation Web App
Protein-Ligand Binding Web App
The fraction of protein with ligand-bound is a function of protein concentration, ligand concentration, and binding affinity (Kd), not just the molar ratio between the protein and ligand. This Web app will help you better understand the relationship between these 4 variables.
Multi-protein Ligand Binding
MBIR
A cryo-ET 3D reconstruction method that effectively minimizes missing wedge artifacts and restores missing information. Publication | Software
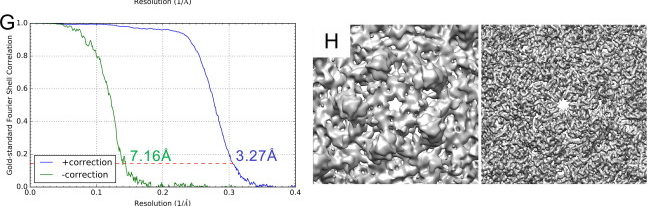
Elliptic distortion estimation and correction
Determine the distortion parameters directly from particle images as part of 2-D alignment without the need of pre-calibration of the microscope. Publication | Available in jspr
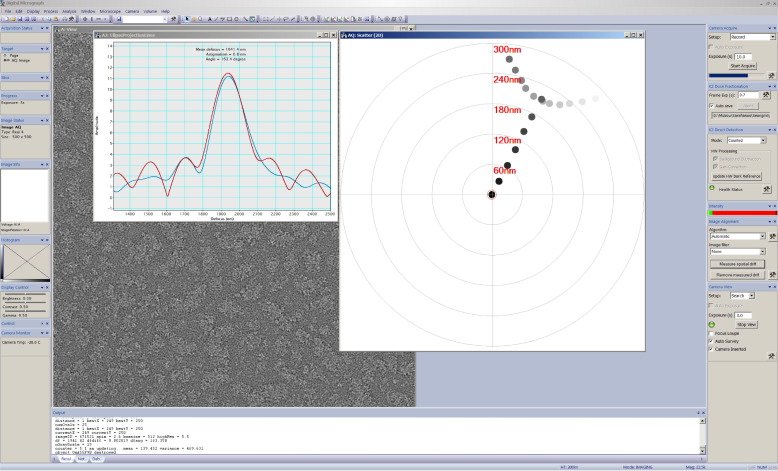
s2stigmator
Real-time detection and single-pass minimization of TEM objective lens astigmatism. Publication | Software
Older Software
- tomoAlign: A fast cross-validation method for alignment of electron tomography images based on Beer-Lambert law. Publication | Software
- tomoThickness: Simultaneous determination of sample thickness, tilt, and electron mean free path using tomographic tilt images based on Beer-Lambert law. Publication | Software
- Motion Correction: UCSF MotionCorr enhanced with rolling window average, internal vertical flip, and support of more image formats. Publication | Software
- FitCTF2: determination of CTF parameters using 3 different methods (s1psfit, s2psfit, s2focus) for cross-validation. Publication | Software | Also available in jspr
- MaskGold and SimGold: Single-Particle Cryo-EM and 3D Reconstruction of Hybrid Nanoparticles with Electron-Dense Components. Publication | Software
- DiaGrid: Single Particle Cryo-EM 3-D Reconstruction: cloud version of jspr. User Guide
- nikontiff2mrc: convert TIFF output image by Nikon SuperCool Scanner to MRC format and convert the pixel values from transmittance to o.d.
- Asymmetric Reconstruction: 3-D reconstruction of complex systems with symmetry mismatches. Download. Original version as part of EMAN software
Software Developed at NCMI@Baylor
- FitCTF: automated fitting of contrast transfer function parameters using a complete EMAN CTF model. Publication | Download
- Icosahedral Virus Reconstruction in EMAN: improved supports for 2-D alignments and 3-D reconstruction of large icosahedral viruses. Distributed as part of EMAN software
- MPSA: improved cross common line method for 2-D alignment of icosahedral viruses. Distributed as part of EMAN software
- FRM2D: 2-D alignment with fast rotation matching method. Distributed as part of EMAN software
- Bilateral Filter: edge-preserving method for denoising 2-D images and 3-D maps. Distributed as part of EMAN software
- SAVR: semi-automated virus reconstruction using common lines and Fourier Bessel reconstruction. Publication | Download
- Helixhunter: identify helices in density maps at subnanometer resolutions and search for homologous protein folds. Distributed as part of EMAN software
- Foldhunter: automated fitting density maps. Distributed as part of EMAN software
- Web CTF: Web-based simulation of the contrast transfer function and envelope functions. Publication | GitHub Archive











Follow Us